Cell fate decisions and organization in the early mammalian embryo
|
In the Posfai lab, we study the molecular mechanisms by which cells acquire their distinct fates during development. We further seek to understand how cells communicate with each other and with their environment to reproducibly pattern an embryo with the correct proportions and spatial orientations of different cell types.
We use the preimplantation mouse embryo as an in vivo model system to study these questions in a physiological setting. The preimplantation mouse embryo is an advantageous model due to its small size, relatively low complexity, and ease of experimental manipulation. In addition to understanding how cellular states are controlled in the embryo, we also aim to stably reproduce these states in vitro, using novel embryo-derived stem cell types. |
|
Preimplantation development in the mouse
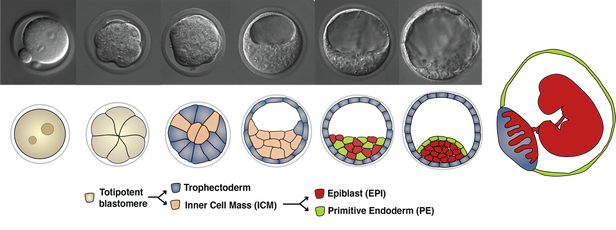
Three cell types, the epiblast (EPI), the trophectoderm (TE) and the primitive endoderm (PE) arise from two consecutive decisions, and self-organize to construct the blastocyst. EPI cells will give rise to the fetus, while TE and PE cells will make extraembryonic tissues, such as the placenta and the yolk sac endoderm, respectively, to support fetal development following implantation. |
- Exploiting our recently developed genome editing method, termed 2C-HR-CRISPR (Gu and Posfai, 2018 Nature Biotechnology) we engineer the embryo genome and create genetically encoded tools to visualize (e.g. fluorescent protein or RNA reporters) and manipulate molecular events (e.g. using protein degron systems, or optogenetic constructs and other synthetic biology tools).
- Using light sheet microscopy, we query cell fate decision making and morphogenesis in live embryos, which allows us to quantitatively address the spatial and temporal dynamics of molecular and cellular features in vivo.
- We develop machine learning-based automated image analysis tools to extract quantitative features from large live imaging datasets suitable for statistical analysis.
|
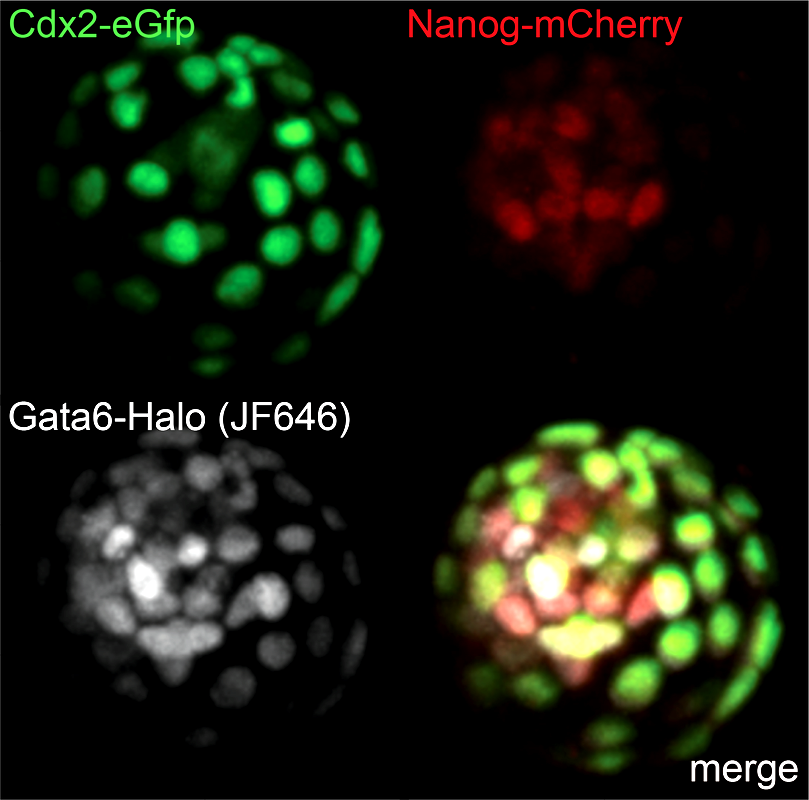
Live triple-color reporter blastocyst, in which all three cell lineages are differentially visualized by tagging endogenous transcription factors with various reporters (Cdx2 - trophectoderm; Nanog - epiblast; Gata6 - primitive endoderm)
|
|
Our most recent collaboration with Stas Shvartsman’s group to establish a robust 3D nuclear instance segmentation for time lapse datasets of developing preimplantation mouse embryos. Check out the preprint on bioRxiv!
|